About
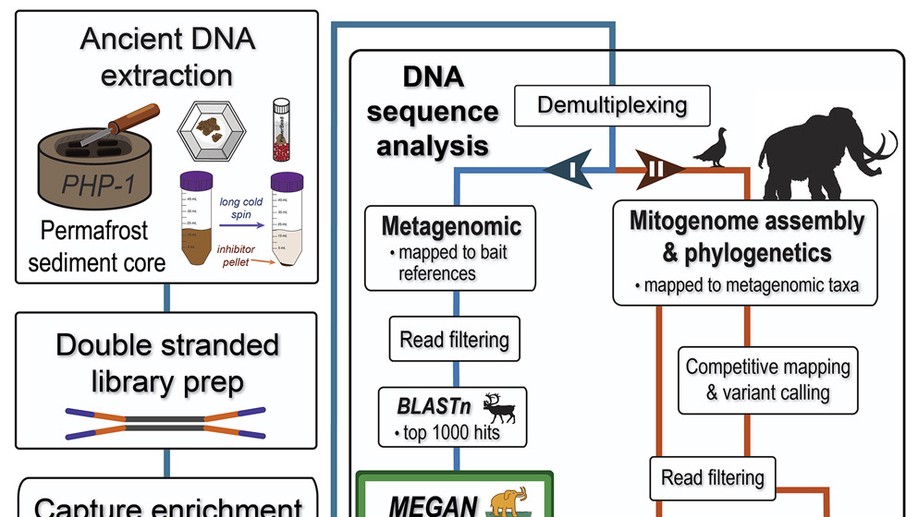
Katherine Eaton is a PhD candidate at McMaster University and she studies the infectious disease “The Plague”. Her dissertation focuses on reconstructing the spread of this disease across the globe, using clinical samples and ancient DNA recovered from archaeological victims of ancient outbreaks.
By investigating past and present incidents of the plague, her work contributes to a better understanding of which populations were affected, why it went extinct in certain geographic regions, and how it has managed to persist throughout human history.
Interests
- Anthropology
- Bioinformatics
- Infectious Disease
- Digital Humanities
- Software Development
Education
-
PhD in Anthropology, 2022
McMaster University
-
BA in Anthropology, 2009
University of Alberta