Pleistocene mitogenomes reconstructed from the environmental DNA of permafrost sediments
Abstract
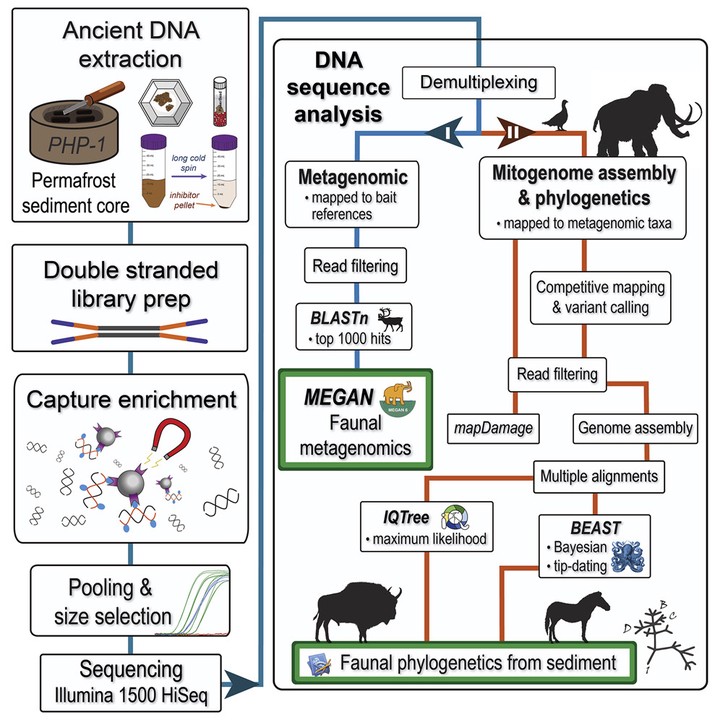
Traditionally, paleontologists have relied on the morphological features of bones and teeth to reconstruct the evolutionary relationships of extinct animals. In recent decades, the analysis of ancient DNA recovered from macrofossils has provided a powerful means to evaluate these hypotheses and develop novel phylogenetic models. Although a great deal of life history data can be extracted from bones, their scarcity and associated biases limit their information potential. The paleontological record of Beringia—the unglaciated areas and former land bridge between northeast Eurasia and northwest North America—is relatively robust thanks to its perennially frozen ground favoring fossil preservation. However, even here, the macrofossil record is significantly lacking in small-bodied fauna (e.g., rodents and birds), whereas questions related to migration and extirpation, even among well-studied taxa, remain crudely resolved. The growing sophistication of ancient environmental DNA (eDNA) methods have allowed for the identification of species within terrestrial/aquatic ecosystems in paleodietary reconstructions and facilitated genomic reconstructions from cave contexts. Murchie et al. used a capture enrichment approach to sequence a diverse range of faunal and floral DNA from permafrost silts deposited during the Pleistocene-Holocene transition. Here, we expand on their work with the mitogenomic assembly and phylogenetic placement of Equus caballus (caballine horse), Bison priscus (steppe bison), Mammuthus primigenius (woolly mammoth), and Lagopus lagopus (willow ptarmigan) eDNA from multiple permafrost cores spanning the last 40,000 years. We identify a diverse metagenomic spectra of Pleistocene fauna and identify the eDNA co-occurrence of distinct Eurasian and American mitogenomic lineages.