Big Data, Small Microbes: Genomic analysis of the plague bacterium Yersinia pestis
In this thesis, I focus on how DNA from the plague bacterium can be used to estimate where and when this disease appeared in the past. To do so, I reconstruct the evolutionary relationships between modern and ancient strains of plague, using publicly available data and new DNA sequences retrieved from the skeletal remains of plague victims in Denmark.
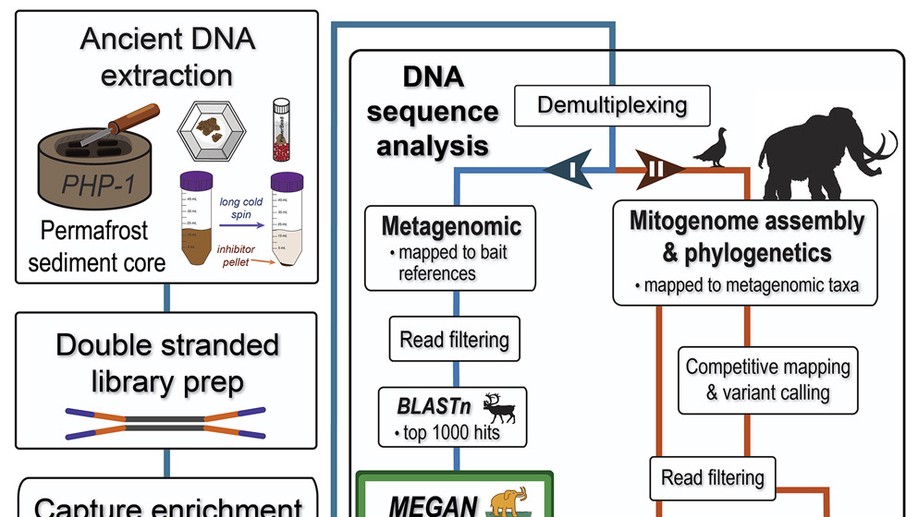
Pleistocene mitogenomes reconstructed from the environmental DNA of permafrost sediments
Murchie et al. used a capture enrichment approach to sequence a diverse range of faunal and floral DNA from permafrost silts deposited during the Pleistocene-Holocene transition.
A Black Death mass grave at Thornton Abbey: the discovery and examination of a fourteenth-century rural catastrophe
This site represents the first Black Death mass grave found in Britain in a non-urban context, and provides unique evidence for the devastating impact of this epidemic on a small rural community.
Genetic resiliency and the Black Death: No apparent loss of mitogenomic diversity due to the Black Death in medieval London and Denmark
This study uses ancient DNA (aDNA) isolated from skeletal remains to examine whether evidence for large‐scale population movement can be gleaned from the complete mitochondrial genomes of 264 medieval individuals from England (London) and Denmark.
Ancient Roman mitochondrial genomes and isotopes reveal relationships and geographic origins at the local and pan-Mediterranean scales
In this paper, we present the whole mitochondrial (mtDNA) genomes of 30 Roman period (1st–4th centuries CE) individuals buried in the Vagnari necropolis in southern Italy. We integrate the mtDNA data with previously published bioarchaeological and isotope (δ18O and 87Sr/86Sr) data for the Vagnari assemblage and compare Roman haplogroup composition to 15 newly sequenced mitochondrial genomes obtained from a pre-Roman Iron Age skeletal assemblage, located in close proximity to Vagnari.
'Written in Bone': New Discoveries about the Lives and Burials of Four Roman Londoners
The Museum of London selected four individuals for multidisciplinary scientific analyses in order to establish their ancestry, aspects of their personal appearance and health. We also reinterpreted their burial context in order to better understand how identity was constructed and expressed in this unique Roman settlement.
Museum of London Report on the DNA Analyses of Four Roman Individuals
Technical report on the ancient DNA analyses of four Roman individuals from London.